
Introduction
The identification of human remains often relies on DNA samples extracted from bones and teeth, which can pose significant challenges due to their highly organized and specialized connective tissue structure, as well as the extensive mineralization present. Despite the advantages of having a better-conserved DNA in these samples, extracting and analyzing the DNA from bones and teeth is significantly more difficult than from fresh tissues due to low endogenous DNA levels, environmental factors, bacterial and postmortem DNA damage, and the presence of co-extracted inhibitors.
Forensic analysts face problems such as significant DNA degradation, low template DNA, and suppressed genetic material, which limit the development of STR profiles for identifying reasons. The complexity of these challenges means that selecting an efficient DNA extraction process is critical to avoid excessive mineral sampling and remove PCR inhibitors. However, there are now commercial technologies available, such as the BcMag™ Bone and Teeth DNA Purification Kit, that offer a solution to these issues.
BcMag™ Bone and Teeth DNA Purification Kit are designed to extract total nucleic acids from bone and teeth samples efficiently and sequentially. The kit uses our unique proprietary magnetic beads in combination with an optimized demineralization buffer to higher yield and super quality of DNA. Purified genomic DNA has the highest integrity and can be used in various downstream applications such as qPCR, STR, etc. The procedure employs mild lysis conditions, avoiding harsh conditions such as alkaline lysis and toxic chemicals for lysing cells to maintain DNA integrity and the time-consuming cleanup of organic solvent from the sample.
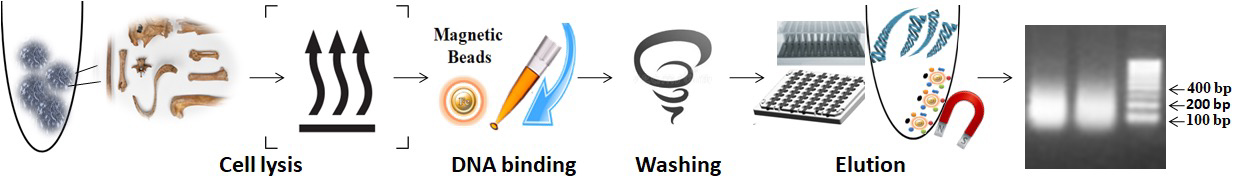
The workflow for Bone-Teeth DNA Isolation is as follows:
1.
Add the lysis buffer and proteinase K to the sample and incubate at 65°C to lyse the bone.
2.
Mix the functional magnetic beads with the sample and vortex or pipette the mixture.
3.
Wash the magnetic beads.
4.
Use a magnet to separate the beads from the sample.
5.
Elute the purified DNA from the magnetic beads.
PROTOCOL
The following protocol is an example. The protocol can be scaled up or down as needed.
Notes
●
DNA yield: Varies (depends on sample size and type)
●
DNA size: Varies (depends on the quality of starting material)
●
For long-term storage, store the extracted nucleic acids at -20°C.
●
Proteinase K preparation: Provide protease K as lyophilized powder and dissolve at a 20 mg/ml concentration in Proteinase K Suspension Buffer. Divide the stock solution into small aliquots and store at -20°C. Each aliquot can be thawed and refrozen several times but should then be discarded.
●
DTT solution preparation: Provide DTT as powder and dissolve at a concentration of 1M in dH2O. For example, 77 mg dissolved in 500µl dH2O. It is stable for years at -20°C. Prepare in small aliquots, thaw it on ice, and use and discard. Store them in the dark (wrapped in aluminum foil) at -20°C. Do not autoclave DTT or solutions containing it. Avoid multiple freeze-thaw cycles.
A. Materials Required by the User
●
95–100% ethanol
●
80% isopropyl alcohol
●
65°C Incubator chamber
●
Microcentrifuge tubes, 1.5ml
●
Aerosol-resistant micropipette tip
●
Magnetic rack: Based on sample volume, the user can choose one of the following Magnetic Racks:
BcMag™ Rack-2 for holding two individual 1.5 ml centrifuge tubes (Bioclone, Cat. No. MS-01)
BcMag™ Rack-6 for holding six individual 1.5 ml centrifuge tubes (Bioclone, Cat. No. MS-02)
BcMag™ Rack-24 for holding twenty-four individual 1.5-2.0 ml centrifuge tubes (Bioclone, Cat. No. MS-03)
BcMag™Rack-50 for holding one 50 ml centrifuge tube, one 15 ml centrifuge tube, and four individual 1.5 ml centrifuge tubes (Bioclone, Cat. No. MS-04)
B. Sample collection
The quality of an STR profile derived from a bone sample is determined by the bone’s type, age, and environmental storage state. The quality of DNA is greatly influenced by soil and humidity conditions. The success of purifying nuclear DNA from bone is also dependent on DNA integrity.
Finding good sampling locations in bones is exceptionally problematic. When working with cremated remains, materials must be handled to reduce contaminants while maintaining adequate material for DNA extraction. Incorrect extraction handling and sample storage might result in cross-contamination of target DNA with non-target DNA. It has been proposed that bone density influences DNA yield because denser tissues give higher physical protection from harm. As a result, DNA is frequently extracted from teeth protected by strong enamel or dense, weight-bearing long bones (tibia or femur). The petrous (or petrosal) part of the temporal bone is denser than many other skeletal sites and has been demonstrated to give much more usable DNA, sometimes four to sixteen times more than teeth.
To effectively extract DNA from the calcium matrix, bone must be preprocessed by removing and discarding the bone or teeth surfaces using scalpels and forceps. The extraction process’s success depends on the degree of grinding, which can be performed through physical grinding or using a low-speed drill to reduce heat buildup. The extraction procedure works best with finely ground bone because cells dispersed in the bone matrix are more accessible for lysis.
C. Purification
1.
Add 10mg of pulverized bone powder into 1.5ml tubes.
2.
Make bone lysis cocktail according to the instructions below, allowing for an excess of n+2 samples:
Components
1x Lysis Buffer
Proteinase K (20mg/ml)
1 M DTT
Total
100 μL Reaction Volume for 5 mg Bone
85 μl5 μl10 μl100 μlComponents
1x Lysis Buffer100 μL Reaction Volume for 5 mg Bone85 μLProteinase K (20mg/ml)100 μL Reaction Volume for 5 mg Bone
5 μL1 M DTT100 μL Reaction Volume for 5 mg Bone
10 μLTotal100 μL Reaction Volume for 5 mg Bone
100 μLPrevious slideNext slide
3.
Add 100 μL of lysis cocktail to each 1.5ml tube containing the bone powder.
4.
Mix the sample by pipetting.
5.
Incubate the tubes in the Incubator chamber with a gentle shake at 65°C for 24 hours.
6.
Remove the tubes from the chamber and mix by vortex or pipetting.
7.
Centrifuge the tubes at 12,000 × g for 5 minutes.
8.
Carefully transfer supernatant to a new 1.5ml centrifuge tube.
9.
Transfer 20 μL of supernatant to a new 1.5ml centrifuge tube.
10.
Add 97 μL of 80% isopropyl alcohol and mix by vortex or pipetting.
11.
Add 15 μLof BcMag™ HO-DNA Beads and mix by vortex or pipetting.
Note:
●
Vigorously shake the bottle until the magnetic beads become homogeneous before dispensing. Do not allow the beads to sit for more than 2 minutes before dispensing. Resuspend the magnetic beads every 2 minutes.
12.
Incubate at room temperature for 15 minutes with gentle rotation.
13.
Place the tube on the magnetic Rack for 1-3 minutes. Remove the supernatant while the tube remains on the Rack. Add 200μL of 85% Ethanol and mix by pipetting 10-15 times to wash the beads. Place the tube on the magnetic Rack for 1-3 minutes and remove the supernatant completely while the tube remains on the Rack.
14.
Repeat step (13) twice.
15.
Remove the tube from the magnetic Rack and let the beads air dry for 10-30 minutes to evaporate the ethanol completely.
16.
Add 15μL to 30μL of 1x Elution buffer and mix by pipetting 30 times to elute the DNA from the beads. Place the tube on the magnetic Rack for 1-3 minutes and transfer the supernatant to a new centrifuge tube.
17.
The eluted DNA should be stored at -20°C.
D. Troubleshooting
Problem
Low DNA Recovery
Probable Cause
Poor starting sample material
Suggestion
- Use better quality of the sample.
- Add more samples
Problem
Low DNA Recovery
Probable Cause
Poor starting sample material
Suggestion
- Use better quality of the sample.
- Add more samples
Learn More
Instruction Manual
MSDS
DNA & RNA Purification Related Products →


